Our lab is interested in quantitative understanding of the molecular mechanisms underlying different layers of gene regulation.
We aim to address three major questions:
- how different gene regulatory processes are mediated by the interaction between cis-elements and trans-factors;
- how the regulatory defect could cause human diseases (e.g. Cancer);
- how gene regulatory network rewired during evolution.
After transcription, mRNAs undergo a series of intertwining processes to be finally translated into functional proteins. To meet the demands of complex organism development and the appropriate response to environmental stimuli, every step in these processes needs to be finely regulated. Dysregulation could result in pathological conditions. Moreover, changes in these regulatory processes represent the major driving forces underlying the evolution of phenotypic differences across different species. Our lab is interested in quantitative understanding the molecular mechanism underlying different layers of gene regulation.
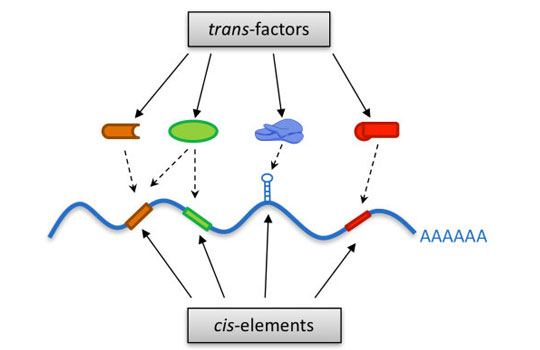
Cis/Trans Gene Regulation
The regulation of different layers of gene expression, including transcription, splicing, polyadenylation, RNA degradation and mRNA translation, are in general all mediated by the interaction between the cis-elements located in DNA/RNA sequences and the diffusible DNA/RNA binding proteins, although the different processes could involve different sets of cis/trans-factors. Mechanistic understanding of the global cis/trans regulatory network demands systematic characterization of the involved cis/trans-components.
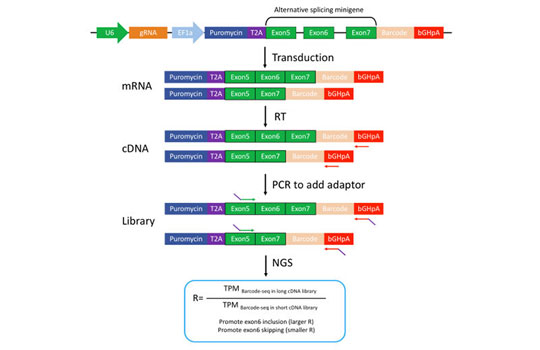
To address this, our lab has been developing novel high-throughput screening strategies to systematically search for cis-elements and trans-factors involved in different regulatory processes.
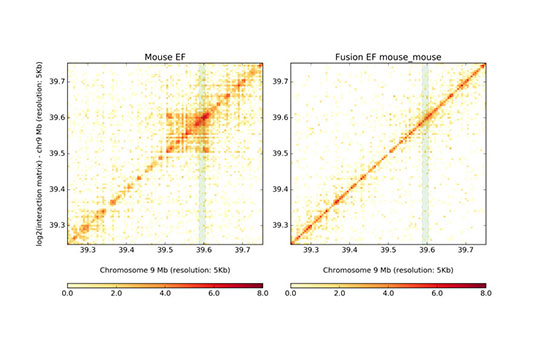
Chromatin Architecture and Gene Regulation
Understanding how chromatin is organized within the nucleus and how this 3D architecture influences gene expression, cell fate decisions and diseases are important questions in the field of gene regulation. Our lab is particularly interested in the following aspects: 1) dynamic changes of chromatin organization during disease development; 2) contribution of cis-elements to allelic differences in chromatin organization and gene regulation; 3)the DNA/RNA binding proteins involved in chromatin organization.
Evolution of Gene Regulation
Complex species exhibit more extensive regulation of gene expression than simpler organisms, and regulatory changes often underlie phenotypic differences between closely related species such as chimpanzee and human.
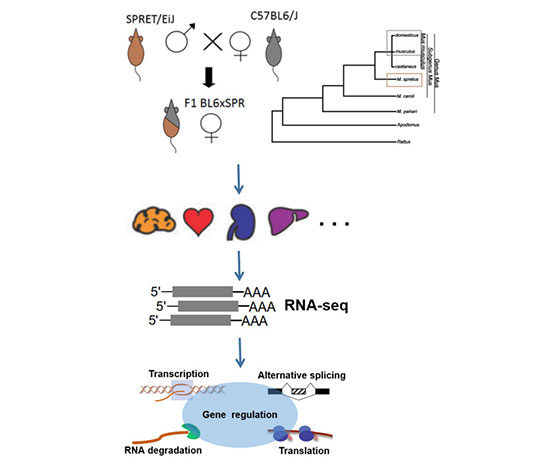
In our lab we study the cis- and trans-regulatory changes underlying differences in the processes of transcription, polyadenylation, splicing, mRNA decay and translation between two mouse species. We use an F1hybrid to address the following questions:
1. Which cases of variation in different gene regulatory processes are of functional importance, e.g. in the division of labor between different tissues or cell types?
2. How do purifying selection, adaptation and random drift shape the evolutionary patterns of different gene regulatory layers?
3. Which gene regulatory changes contribute to organismal differences and to reproductive isolation between species?
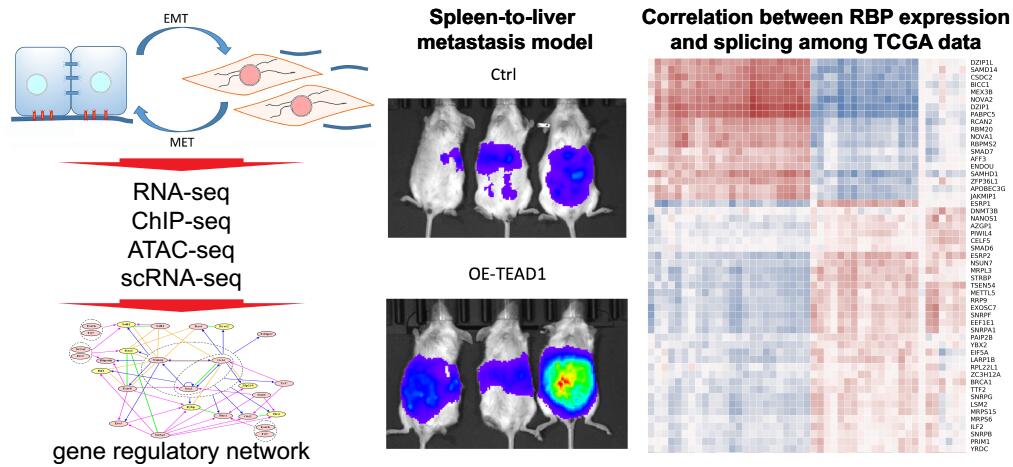
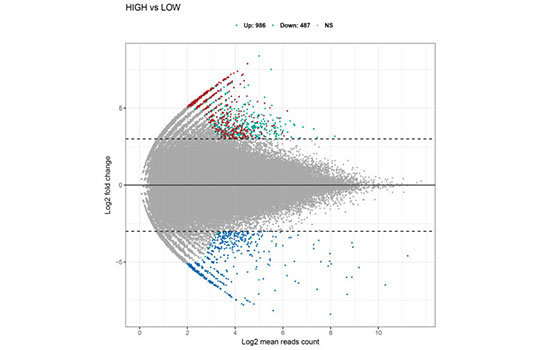
Gene Regulation in Cancer
Epithelial-mesenchymal transition (EMT) is a vital process for large-scale cell movement to initiate the metastasis of cancer cells.
By comparing EMT and non-EMT cells in an epigenetically heterogeneous cell population derived from an isogenic cell line, we have discovered several critical TFs that are transcriptionally or posttranscriptionally regulated to support the EMT process. We are currently studying their function and upstream regulatory mechanisms.
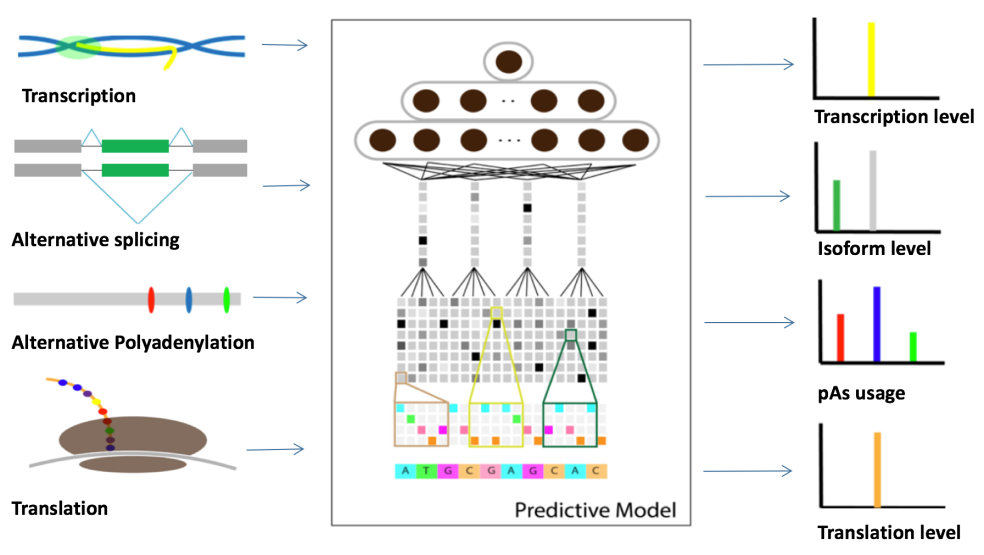
Deep Learning-based Model for Gene Regulation
Recently, deep learning has been reemerging as the driving force of artificial intelligence and machine learning.
Compared to the traditional machine learning algorithms, deep learning is a wrapper method, which trains both the feature extractor and the classifier simultaneously. In recent years, with the significant improvement of computational power and advancement of big data, deep learning has achieved great success in various fields, including genomic studies.
In collaboration with Prof. Xin Gao at KAUST, our lab is developing novel deep learning-based methods to study the regulatory network of gene expression, including transcription, alternative splicing, alternative polyadenylation (APA), and translation, and applying our model in understanding their dysregulation in human diseases such as cancer.
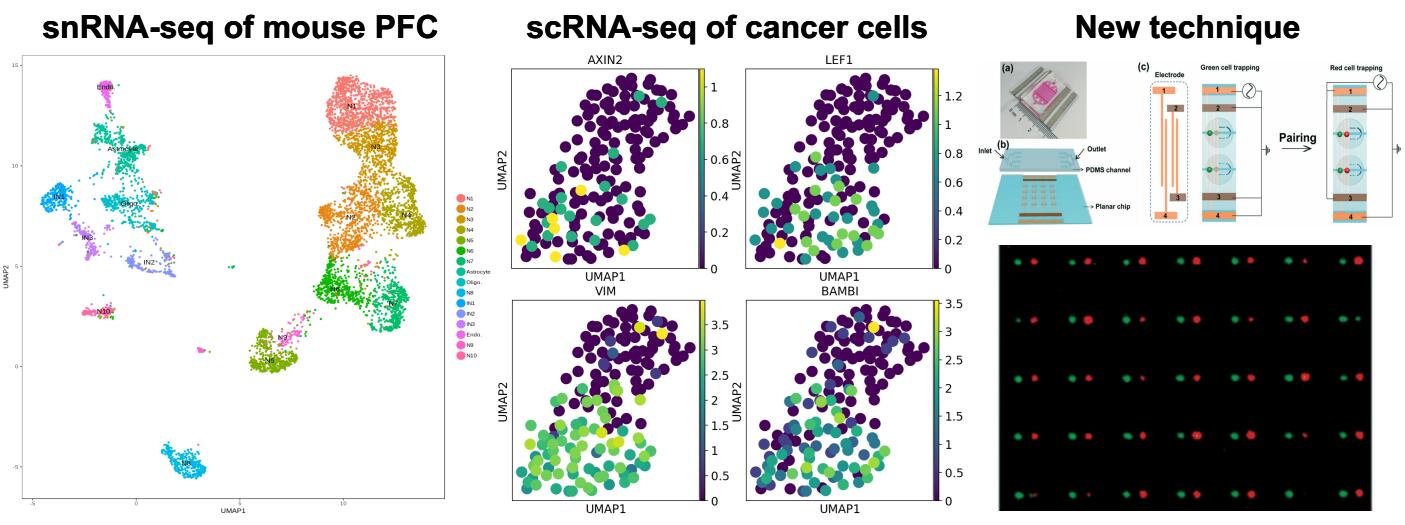
Single-cell Sequencing Technology
Single-cell sequencing methods have revealed new biology in terms of cell composition of tissues, the dynamics of transcription, and the regulatory relationships between genes. Rapid technological developments at the level of cell capture, phenotyping, molecular biology, and bioinformatics promise an exciting future with numerous biological and medical applications.
Our lab is not only using available single cell sequencing techniques to study the heterogeneity of brain and tumor tissues, but also developing novel methods for single cell sequencing in collaboration with Prof. Xin Cheng at the Department of Materials Science and Engineering.
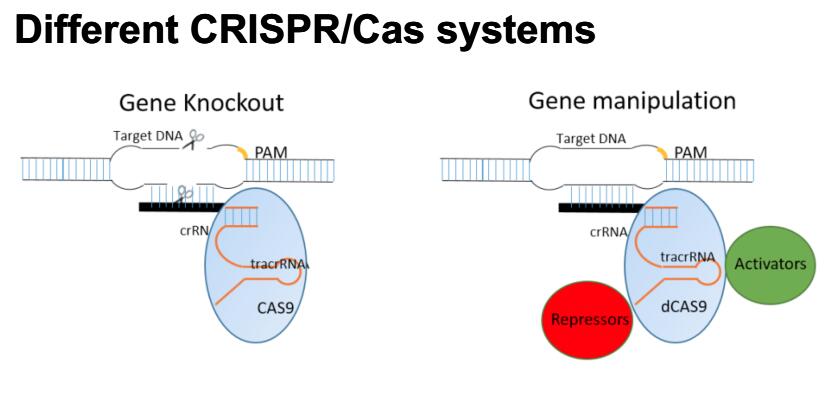
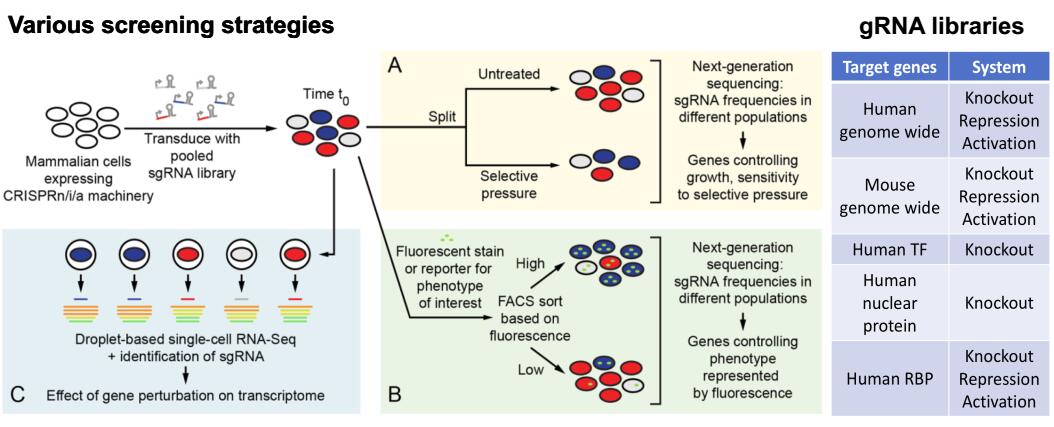
CRISPR/Cas-based Genetic Screens
Based on cell survival/proliferation, fluorescent reporters or single-cell RNA-seq, we are combining different types of CRISPR/Cas-based screen systems (i.e. knockout, inhibition and activation)to identify essential genes in cancer cells, genes involved in certain cellular phenotypes and signaling pathways, and novel trans-factors involved in post-transcriptional regulation.